
Dr.
E-mail: m.katika@utwente.nl
Telephone: +31 53 489 2456
Fax: +31 53 489 3511
Address: Faculty of Science and Technology
Medical Cell BioPhysics (MCBP)
Building: Carré
Room: CR4429
Drienerlolaan 5
P.O. Box 217
7500 AE Enschede
The Netherlands
Project goal
The main objective of my work is to isolate and characterize circulating tumor cells (CTCs) in prostate and lung cancer patients using VyCAP's Self-Sorting Nanowell chips
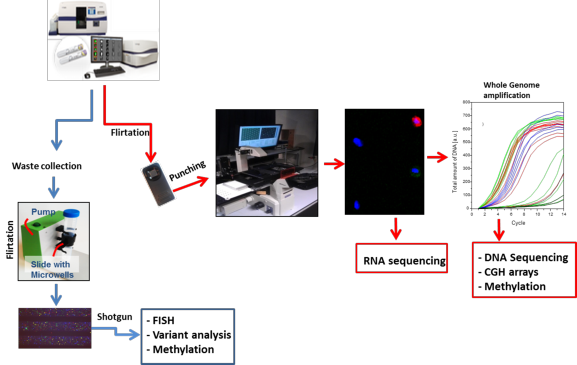
Figure.1 : Schematic illustration of strategies used for isolation and characterization of CTCs.
Project background
The current cancer diagnosis procedures has major challenges at tumor diagnosis including a) need for a tumor biopsy which is difficult to obtain each time for therapeutic intervention b) limited predictive value of current molecular histopathological classifications, therefore cancer patients end up being over-treated or under-treated. There is a great need for development of non-invasive biomarkers to avoid severe pain to the patient and accurate prediction of the tumor progression to improve the selection of patients for right treatment. Isolation of CTCs will offers unique opportunity to develop ‘liquid biopsy’ tests that would enable non-invasive diagnosis of cancer for instance by a simple blood test. CTCs are rare cells that shed into the circulatory system from a primary tumor and circulate in the bloodstream. CTCs can seed on distant organs and initiate growth of new tumors (metastasis). CTCs count represents a promising prognostic marker which might be of value for guiding treatment of patients and predicting cancer. Molecular characterization of CTCs will unravel the molecular mechanisms underlying metastasis, tumor reoccurrence and drug resistance.
Cancer is a heterogeneous disease with several subclasses showing differences in tumor biology, histopathology and prognosis. Due to tumor heterogeneity most cancer treatments are in effective and works in only a minority of patients. The standard pathological examination of tumors often relies on needle biopsy, a procedure in which small samples of cells are extracted from a tumor for analysis. However, the tiny fraction of cells in the biopsy may not be representative of the entire tumor mass which results in lack of important disease features. The heterogeneity of tumor cells dictates the need for gene expression analysis at the single cell level. The isolation of individual CTCs is remains a challenge despite the continued development of new technologies. To that end a variety of technologies including flow cytometry, micromanipulator, laser-capture microdissection and DEPP arrays have been widely used but all have several limitations in terms of efficiency, cost and processing time. We introduce a simple method VyCAP micro well chips for isolation and characterization of individual CTCs. VyCAP microwell bottom is made up of silicon nitride (SiNi) membrane with a thickness of 1 μm, having a single pore with a diameter of 5 μm in the center.
Methods
Spiked and patients samples are subjected to auto prep (Cell search) and cartridges are scanned for CTC enumeration. A cell suspension enriched for CTC is placed on a microwell plates (VyCAP cups). After drying, cells along with the bottom of the well are punched with a needle attached to a microscope stage into a 384 or 96 well PCR plate. The presence of the punched cells in the 384 well plate was verified using a fluorescence microscope. Whole genome amplification kits are used to amplify DNA from punched single cells and subjected to further analysis such as DNA methylation, copy number alterations (CGH arrays) and variant analysis (Targeted and whole exome sequencing). START2 method is used to amplify RNA from single or pooled punched cells for RNA sequencing. Next to this, we are using multiple strategies such as additional filtration (VyCAP sieves) and staining methods (cytokeratin’s antibodies) to catch a fraction of CTCs (EpCAM weak or negative) in cell search waste which are not captured in auto prep. We punch those CTCs in to 96 well plates using shotgun method and are further investigated for tumor characteristics (e.g. chromosomal aberrations, mutations and methylation).
Outcome
This study enables the identification non-invasive biomarkers and novel targets for development of personalized medicines.
