Research in the CRN Lab
We investigate how Chemical reaction networks (CRNs) can become capable of functions akin artificial intelligence. To this end, we work on three synergistic research lines in the CRN lab, focusing on how CRNs can i) memorize, ii) communicate, and iii) self-organize. In greater detail, we aim to; |
i) | understand the driving force that enables CRNs to temporarily memorize past events, a prerequisite for (artificial) intelligence. As a starting point we demonstrated how gradients drive an enzymically-driven autocatalytic network to display history-dependent behaviors. |
ii) | establish a platform for local biochemical feedback in CRNs. We recently demonstrated how reactive surfaces (that built on polylysine-coated surfaces) enable CRNs with the capacity of signal transduction. We also explore if such systems could provide the foundation for systems with biosensing capabilities. |
iii) | use hypergraphs to generalize how reactions self-organize into CRNs. We are currently developing a novel mathematical framework that translates synthetic CRNs into graphs. This work builds on my expertise in the synthesis of out-of-equilibrium networks and collaboration with Dr. Stegehuis (expert in graph analyses). |
Temporal and local control over feedback systems
|
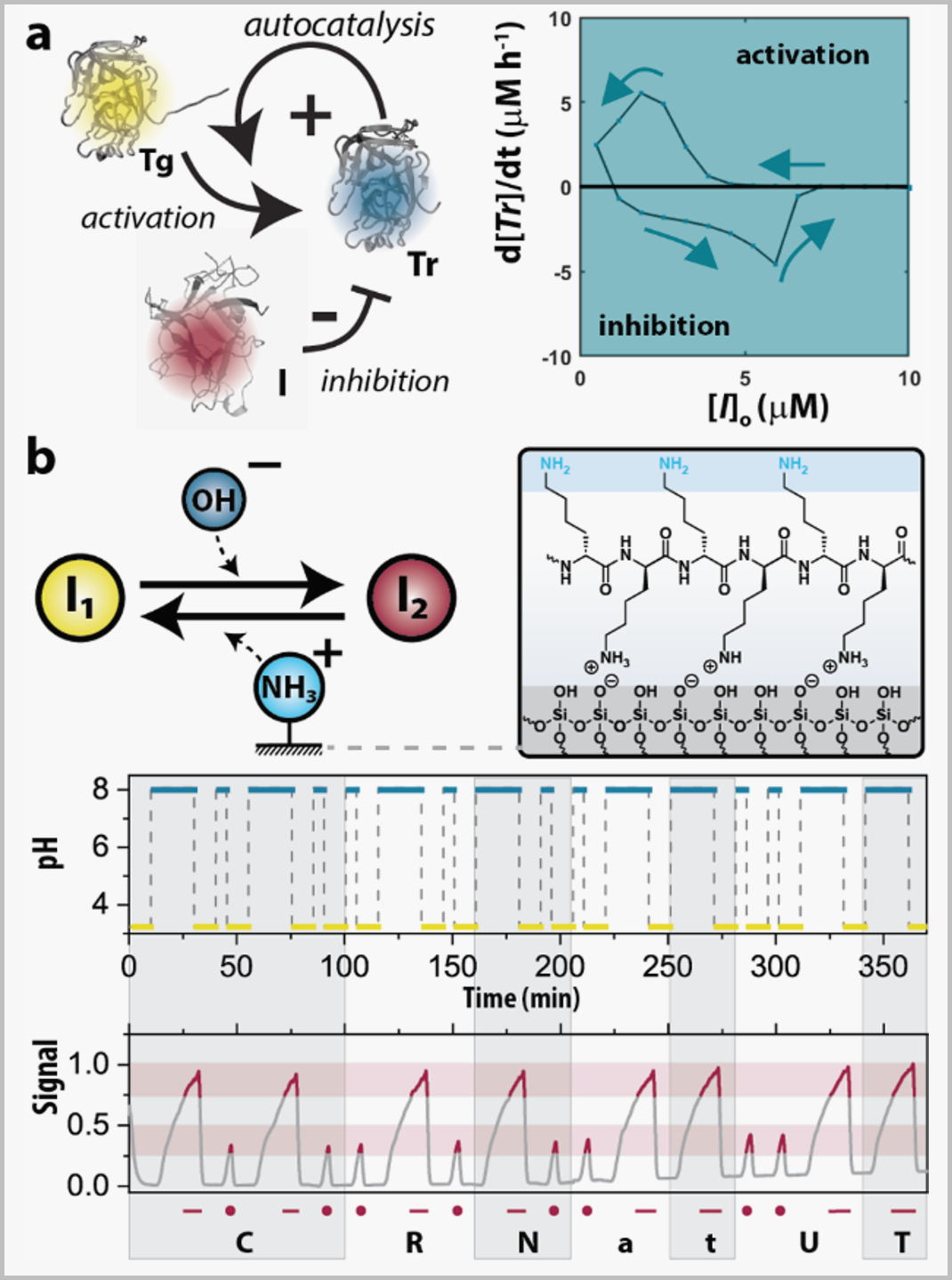
Fig. Examples of chemical systems from the CRN lab: A (a) gradient-driven autocatalytic network (#10), and (b) surface-driven competing activation network (#12). |
Methods that enable a balance between (often opposing) feedback loops and maintain out-of-equilibrium conditions are essential in the de novo design of CRNs. We recently demonstrated how gradients drive CRNs to display history-dependent behaviors (Fig. a). Under continuous flow, Tg acts as a fuel for generating the catalyst Tr, creating a positive feedback loop (+), and its process is regulated by the inhibitor, which can suppress the activation of autocatalysis (-). The incorporation of an inhibitor into the network was crucial as it enables a controlled release of Tr, giving rise to behaviors such as hysteresis and adaptation. We, thus, showed that CRNs can temporarily memorize past events (#10), a prerequisite for (artificial) intelligence. In another work, we reported how reactive surfaces could create CRNs with the capacity to transduce, delay, and perturb chemical signals (#12). Fig. b depicts an acid-base equilibrium which can be perturbed by two opposing processes, one of which is immobilized on a surface. Fixating poly-l-lysine (PLL) on the surface allowed us to work under flow so that the PLL can exert its function (in this case, binding/ releasing protons) on the liquid-surface interface. Our demonstrations include write and read operations to encode ‘CRN@UT’ (my at research group at the University of Twente).
background
Intelligent behavior, similar to other complex behaviors, can emerge under out-of-equilibrium conditions. In my first research article, published in Nature Chemistry (2015, #1), I demonstrated that oscillatory behavior (i.e., a hallmark of life, and a phenomenon that can only sustain under out-of-equilibrium conditions) could be rationally designed. Its ground-breaking character is emphasized by the high FCWI (6.5) and my publications in JACS (2015, #2, 2017, #3, 2017, #6, 2019, #7) and Nature (2023, #11) that further laid out the underlying principles. Overall, I established how an integral approach involving chemical synthesis, microfluidic fabrication, and mathematical modelling is required to design chemical systems with complex behaviors (#5). Among other demonstrations, this approach formed the foundation for establishing how competing reactions can become cooperative (#6, #7), which could translate into the design of an autonomous periodic catalyst (#11).