Deep learning for fungal identification based on fungal growth images
Description of the assignment:
Most building materials are of organic origin (e.g. wall- paper and wooden floor) and therefore are prone to fungal growth when the materials are damaged by water e.g. leaking water pipes, flooding or faulty building constructions. Condensation is another indoor problem caused by high humidity in combination with thermal bridges in the building and lack of airing or ventilation often exacerbates these problems. When gypsum tiles and wallboard which are often covered with heavy paper or wall paper become wet, they become one of the best substrates for undesirable fungi, like Chaetomium, Trichoderma and Stachybotrys (Flannigan et al. 2011). It has been reported in the Netherlands’ Housing Survey (WoON) 2018 that 19% of the houses in the Netherlands have been affected by moisture and molds. Fungal growth in water-damaged buildings can have many adverse health effects on the users (WHO, 2009) and prevention is the best method to avoid it.
In the food industry, spoilage of the raw materials or final products may occur and is connected to the development of a specifically adapted mycobiota of various processing sites, storage rooms and air- and other filter systems is advisable. In bakeries, for example, dry conditions prevail, flour and other dried raw materials and in the final products, which favor growth of xerophilic fungi, e.g. Eurotium species and Wallemia sebi. Early detection of these species during the production is essential to prevent soilage and fast remediation.
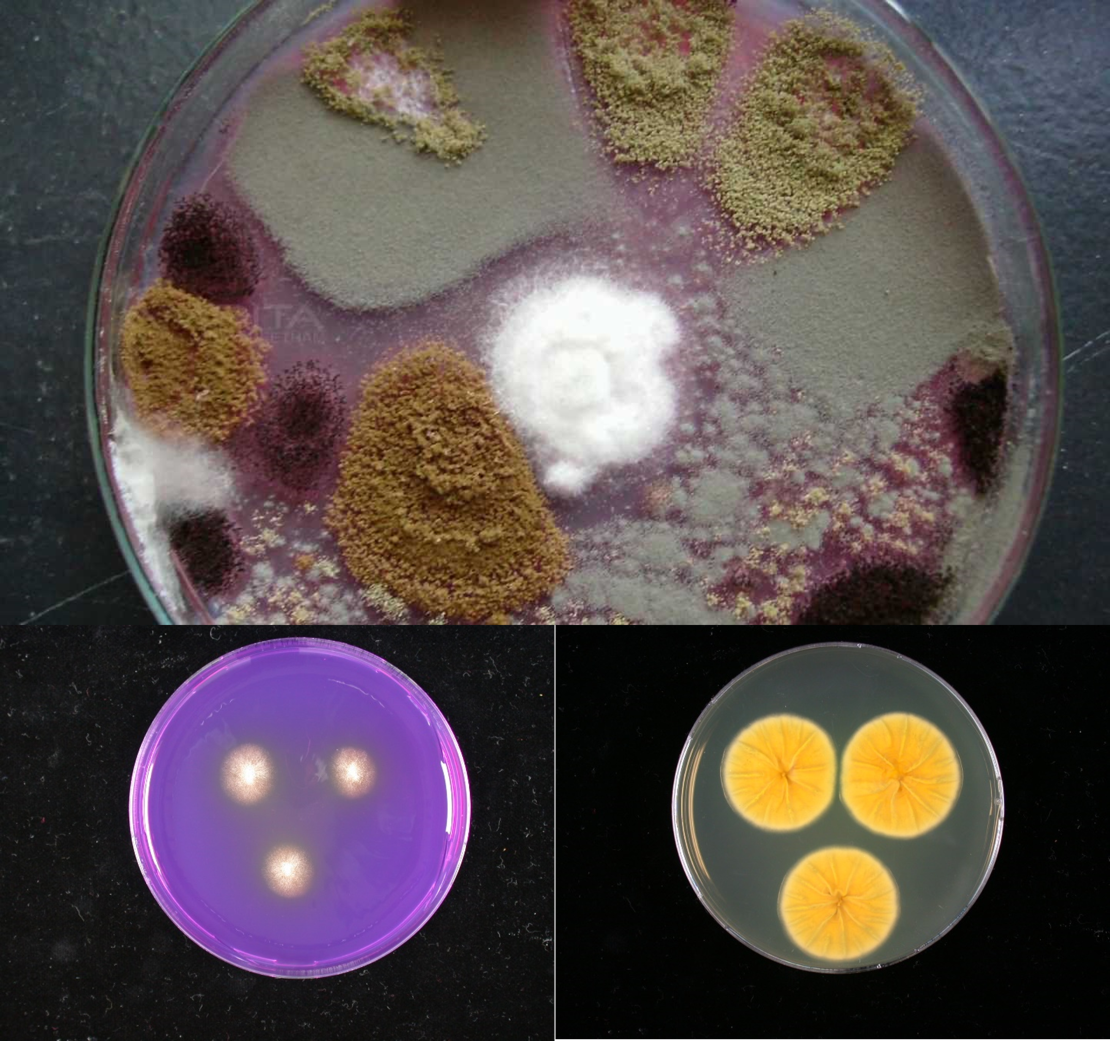
Currently, there are two methods for fungal identification in indoor and food environments. The first method is the morphological identification. Samples collected from contaminant spaces potentially forming “nursery chambers” for fungal growth are cultured on special media recommended by specialists. Fungal cultures grown can be cut out in a square and examined under the microscope. Based on the mycelium, fruit bodies, sporulating structures and the number of the colonies appearing after a given time, the fungi present in the samples can be detected. This method has been proven very useful for examining visible of fungal contamination of building materials or food products. However, it requires experienced staff working with indoor and food fungi. The second method is the molecular identification. The DNA of collected samples are extracted and subsequently genetic markers for fungal identification are sequenced. The obtained DNA sequences are compared with the known sequence databases to find the best matches. A possible species identification can be ascertained based on the percent similarity with the best match. This method is more reproducible and accurate than the morphological identification method. However, the complete procedure takes more time and more expensive. In addition, not all laboratories have access to the sequencing facilities.
At the Westerdijk Fungal Biodiversity Institute, we have a large collection of more than 100000 living strains representing ca. 17000 fungal species, containing most of the indoor and food fungi. In addition, we have a unique set of images of many of the indoor and food fungal growth on various condition media. Over the last few years we have seen enormous success in the field of image analysis and computer vision. In particular, deep learning has emerged as a successful approach for large scale machine learning for images classification. In this proposal, we aim to develop a new method for fungal identification by employing the deep learning approach for classification of fungal growth images. Using a collection of fungal growth images of indoor and food fungi as a training dataset, we hope to detect the source of contamination in the indoor and food environments quickly and accurately.
Originality
At mentioned earlier, at the Westerdijk Fungal Biodiversity Institute, we have an almost complete and unique collection of images of fungal growth of indoor and food fungi. In addition, we have also experienced staff who have a lof of knowledge about these organisms. Deep learning has emerged as a successful approach for large scale machine learning for images classification through all areas of science and industry. It is able to learn a representation of the images reflecting experts’ knowledge. Thus, the learning algorithm does not require manual, handcraft features like in traditional machine learning methods. To the best of our knowledge, this is the first time that deep learning will be applied for fungal identification based on fungal growth images.
Potential societal impact and relevant stakeholder(s)
As the proposed method could easily, quickly, cheaply and accurately identify indoor and food fungi, it has a number of potential applications. First of all, it will be very useful for the laboratories working on fungal identification who have no access to the sequencing facilities. Secondly, in homes or workplaces where people need a quick analysis for fungal contamination. Finally, although, the Westerdijk Fungal Biodiversity Institute, we have access to the sequencing facility, this tool will be useful for us as well as the molecular identification next to the cost, usually takes 20 days for fungal identification of a mixed cultures. Although this tool is made primarily for indoor and food fungi, it can be reused for other fungal organisms. This dataset of fungal growth images have not been used before. A successful project would potentially generate income for the institute.
Contact:
Supervisor at Westerdijk Institute: Duong Vu d.vu@wi.knaw.nl
UTwente supervisor: Jacob Kamminga j.w.kamminga@utwente.nl
