Proteins are molecules of life. They are ubiquitous in living cells as they carry out most of the cell functions. Consequently, nature of most diseases is often related to malfunctioning of the proteins. To name a few examples: Alzheimer’s disease is related to aggregation of malfunctioning proteins in the brain; cancer is often caused by malfunction of the proteins responsible for keeping reproduction of cells under control; pathogens like viruses and bacteria can invade organisms using proteins to penetrate the cell membrane or trick the immune system. Therefore, many analytical disciplines focus on studying and characterizing proteins, their dynamics and interactions.
The past decade witnessed the advent of novel techniques to study proteins at unprecedented single-molecule resolution. For example, the technology of optical tweezers allows to pull on the protein molecule and observe it’s reaction to force, or the FRET(Forster resonant energy transfer) allows to measure distance between different parts of the protein by decorating them with molecules that can emit light(fluorescent labels) and observing their interaction in the microscope. Inception of these revolutionary technologies generated a lot of knowledge about how living systems operate at the molecular level. However, despite the wide variety of techniques available to study proteins, certain protein functions and interactions are still elusive. We are developing a new label-free and specific method that will monitor protein physical properties (scattering cross-section, hydrodynamic radius and permanent dipole moment) through single-molecule manipulation and optical scattering measurements. These properties can report in real time on protein conformational dynamics, interactions with other proteins and ligand binding.
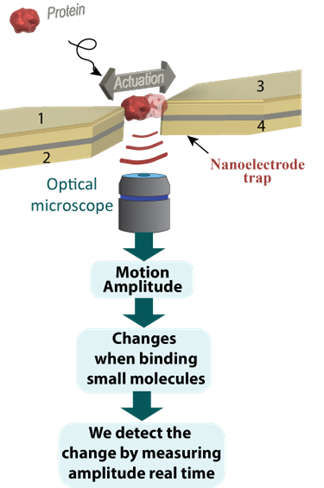
Figure 1. Concept of the experiment.
Our approach is based on a physical manipulation of the protein under study. In a nutshell, we develop a nanoelectrode system (Figure 1), which will allow to capture a protein molecule and engage it in a periodic motion, tossing it from one electrode to another like in a mini game of ping-pong. Since the protein will be actuated by the electric field, its trajectory will depend on the strength of the electric field, size and on the dipole moment of the protein. Monitoring and analyzing the motion amplitude we will be able to discern and monitor protein parameters such as size and dipole moment.
Team of the project:




