The way a virus like Influenza A is transmitted from animals to humans, is still not well understood. How does the virus recognize the host cells in humans, even if the surface is different from that of animals? Researchers of the University of Twente mimicked the cell surface on a molecular scale, including the sugars to which the virus attaches. Binding proves to be complex. The density of binding molecules is different, but the type and length as well. The research, published in ACS Central Science today, shows that a minimum number of bindings is needed for a ‘successful’ transmission.
Viruses bind to host cells using the well-known spike-shaped proteins at their surface. In the case of Influenza A, these proteins are hemagglutinins binding to the sugars at a cell surface, the sialic acids. The virus has to find a way to interact sufficiently strong, before it is encapsulated and starts its destructive job. In essence, in human and animal cells it is the same type of binding. This is, however, not the full explanation, because many types of influenza will never be transmitted to humans. A crucial difference is the number of sialic acids on a surface. The structure and length of the sugar molecules in humans are different as well, resulting in less opportunities for binding. An avian virus does not have a good starting point when it ‘lands’ on a human cell surface: it doesn’t ‘see’ all the necessary bindings. Still, it has to find a minimum of around eight interactions to be effective, the research shows.
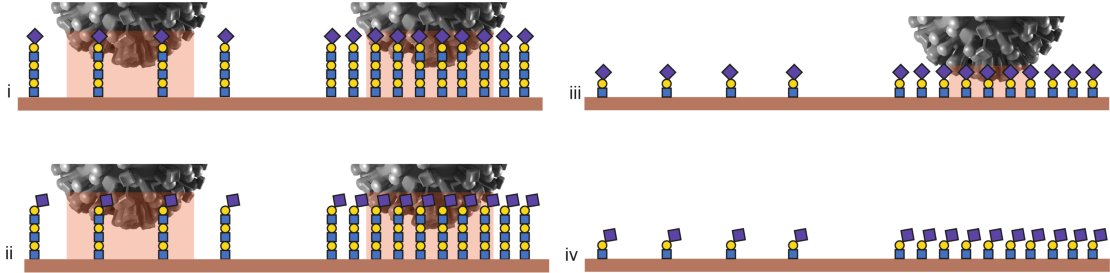
All pictures (i-iv) show a low receptor density on the left and a high density on the right. In (i) and (ii), the virus will bind at both densities, thanks to the length of the molecules. In (iii), the molecules are favourable but have a limited length so the virus will only bind at high densities. In (iv), none of the boundary conditions are met, so the virus does not bind.
Learning more about zoonosis
In order to determine the binding strength, the researchers have mimicked the cell surface with sialic acids on it. They created a gradient in this, ranging from a low density of molecules to a higher density. At low densities, a virus doesn’t have sufficient interaction opportunities. The length and structure of the molecules are decisive as well. The new tool gives valuable insight in the binding opportunities, and the infection risk. Although this research focused on Influenza A, the new insights improve our understanding of other viruses, like corona viruses, as well. So, the tool can be help us understand the probability of zoonosis.
Multidisciplinary team
The research has been done in the Molecular Nanofabrication group (MESA+ Institute) of Prof Jurriaan Huskens. It was done in a multidisciplinary team including virologists (Royal GD veterinary lab), sugar and biology chemists (Utrecht University), and experts of molecular dynamics (University of Georgia) and theoretical/computational physics (TU Eindhoven).
The paper ‘Hierarchichal multivalent effects control influenza host specificity’, by Nico Overeem, Erik Hamming, Oliver Grant, Daniele Di Iorio, Malte Tieke, Candelaria Bertolini, Zeshi Li, Gaël Vos, Robert de Vries, Robert Woods, Nicholas Tito, Geert-Jan Boons, Erhard van der Vries en Jurriaan Huskens is published in ACS Central Science of the American Chemical Society.





