Investigating functional effects of molecular crowding in stem cells
Stem cells are of great interest for tissue engineering application due to their capacity for multi-lineage differentiation. However, the differentiation and tissue formation of stem cells has remained relatively inefficient. Recent breakthroughs have identified that enhanced macromolecular crowding significantly improves these processes. However, the mechanism behind the phenomenon is at best poorly understood. In this project we will use advanced cell/molecular biological approaches to unravel important aspects of this mechanism.
Specifically, the effect of molecular crowding on the activity of master transcription factors for chondrogenic, osteogenic and adipogenic differentiation (SOX9, RUNX2, and PPARg respectively) will be investigated using a novel biophysical microscopy-based technique Transcription-Factor Fluorescence Recovery After Photobleaching (TF-FRAP). Human stem cells will be sorted on cell size and/or cell surface markers using fluorescent activated cell sorting (FACS) and functionally analyzed using qPCR, western blotting/immunofluorescence and TF-FRAP. The student will therefore be trained to become experienced in a wide range of highly relevant (stem) cell biological methods and techniques.
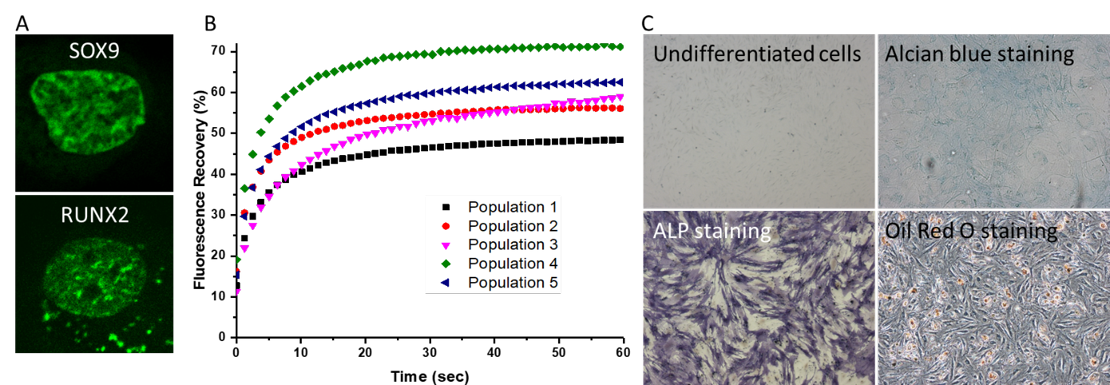
Figure 1. (A) Nucleus expressing SOX9 or RUNX2. (B) TF-FRAP recovery curves show differential mobility of SOX9-mGFP among subpopulations of hMSCs. (C) Alcian blue, ALP and Oil Red O staining show the differentiation of hMSCs to chondro, osteo and adipogenic lineage respectively.
Methods: FACS, TF-FRAP, transfection, plasmid and mRNA isolation, qPCR, hMSC differentiation, staining, western blotting, immunofluorescence.
