In 2019 cervical cancer was the fourth most common malignancy in females, with more than half a million diagnosed cases and over 300.000 deaths worldwide [1]. A substantial amount of women do not participate in screening programs designed to detect and monitor high-risk HPV infection, increasing their risk of developing high-grade cervical intraepithelial neoplasia (pre-malignant) or cervical cancer (malignant) [2]. To make participation more attractive, self-sample tests for high-risk HPV types are offered. This raises the need for a non-morphological biomarker (panel) that can be implemented directly on the self-samples.
Recently, a study by Snoek et al. implicated a panel of 9 miRNAs as a molecular signature for CIN stage 3 or worse, that could be used as a novel triage marker [2]. Micro-ribonucleic acids (miRNAs) are short non-coding RNA molecules involved in the post-transcriptional regulation of gene expression [3]. The validation of the miRNA signature with qPCR was complicated by the apparent presence of so-called isomiRs, which are length variants of miRNAs [4]. It is possible that specific length variants are more clinically relevant than their exact sequences [2]. This raises the need for a detection method capable of discriminating between miRNA isoforms at single base resolution.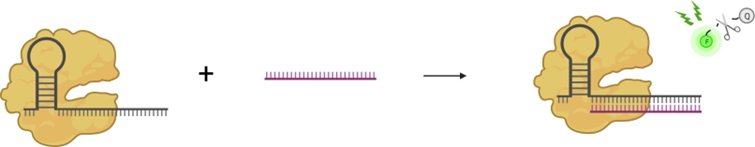
Figure 1: Schematic overview of trans-cleavage of a quenched fluorophore reporter for biosensing purposes.
Due to its high specificity, efficiency, and ease of programming, CRISPR-Cas13a is an alternative candidate for isomiR discrimination [5]. Type VIa CRISPR-Cas employs Cas13a to cleave single-stranded RNA (ssRNA) [7]. First, the Cas13a protein will associate with CRISPR RNA (crRNA). The crRNA is a programmable ssRNA molecule that serves as a cleavage guide for Cas13a [6]. Once the crRNA associates with the complementary target RNA, Cas13a starts cleaving not only the target RNA (cis-cleavage) but all surrounding ssRNA molecules (trans-cleavage). This characteristic is the basis for numerous Cas13a- based nucleic acid detection systems, following the general principle schematically shown in Figure 1.1.
In this assignment, you will work on an isomiR discrimination assay based on strand displacement of a short nucleic acid sequence (Figure 1.2). This assignment will consist of (CRISPR) assay optimization (continuing on the work performed by a former MSc student [8]), nucleic acid design, and finally, applying the assay to real patient samples. After successful development, the assay will also be adapted to a microfluidic chip for easy, reliable, and fast patient diagnosis.
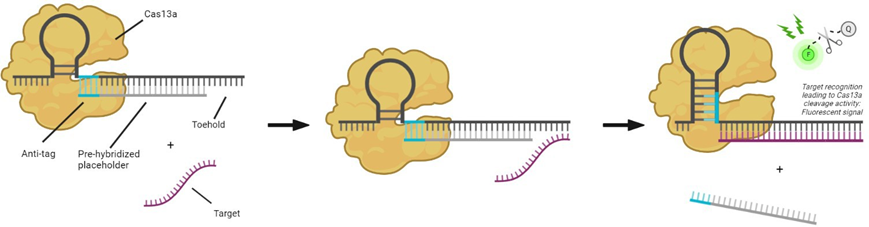
Figure 2: A placeholder with extended complementarity will be pre-hybridized with the crRNA, keeping Cas13a inactive. In the presence of a target RNA with more subsequent complementary bases, the placeholder will be displaced by the target RNA. Cas13a can now be activated.
Contact person:
References:
[1] Paul A. Cohen et al. “Cervical cancer”. In: The Lancet 393.10167 (Jan. 2019), pp. 169–182. ISSN: 0140-6736. DOI: 10.1016/S0140-6736(18)32470-X.
[2] Barbara C. Snoek et al. “Genome-wide microRNA analysis of HPV-positive self-samples yields novel triage mark- ers for early detection of cervical cancer”. In: International Journal of Cancer 144.2 (Jan. 2019), pp. 372–379. ISSN: 1097-0215. DOI: 10.1002/IJC.31855. URL: https://onlinelibrary.wiley.com/doi/ full/10.1002/ijc.31855%20https://onlinelibrary.wiley.com/doi/abs/10.1002/ ijc.31855%20https://onlinelibrary.wiley.com/doi/10.1002/ijc.31855.
[3] Minju Ha and V. Narry Kim. “Regulation of microRNA biogenesis”. In: Nature Reviews Molecular Cell Biology 2014 15:8 15.8 (July 2014), pp. 509–524. ISSN: 1471-0080. DOI: 10.1038/nrm3838. URL: https://www.
nature.com/articles/nrm3838.
[4] Anita Schamberger and Tamás I. Orbán. “3′ IsomiR Species and DNA Contamination Influence Reliable Quan- tification of MicroRNAs by Stem-Loop Quantitative PCR”. In: PLOS ONE 9.8 (Aug. 2014), e106315. ISSN: 1932- 6203. DOI: 10.1371/JOURNAL.PONE.0106315. URL: https://journals.plos.org/plosone/ article?id=10.1371/journal.pone.0106315.
[5] Jeanne E. van Dongen et al. “Point-of-care CRISPR/Cas nucleic acid detection: Recent advances, challenges and opportunities”. In: Biosensors and Bioelectronics 166 (Oct. 2020), p. 112445. ISSN: 0956-5663. DOI: 10. 1016/J.BIOS.2020.112445.
[6] Frank Hille et al. “The Biology of CRISPR-Cas: Backward and Forward”. In: Cell 172.6 (Mar. 2018), pp. 1239– 1259. ISSN: 0092-8674. DOI: 10.1016/J.CELL.2017.11.032.
[7] Mitchell R. OConnell. “Molecular Mechanisms of RNA Targeting by Cas13-containing Type VI CRISPR–Cas Systems”. In: Journal of Molecular Biology 431.1 (Jan. 2019), pp. 66–87. ISSN: 0022-2836. DOI: 10.1016/J. JMB.2018.06.029.
[8] Joyce Rops. “Early detection of cervical cancer via Cas13a mediated microRNA detection” MSc thesis, University of Twente (October 2022)
