Artificial insemination (AI) is used for the veterinary industry worldwide [1]. On industrial farms, approximately 90% of pigs and 80% of dairy cattle is bred using artificial reproductive technologies. To succeed with AI, it is important to assess and select semen of high quality. In conventional methods, computer-assisted sperm analysis (CASA) is used to measure quality parameters of a small ejaculate sample, such as concentration, average motility and average morphology. Though the conventional method is very efficient in the quality assessment, it lacks the capability of selecting the best spermatozoa for AI. In previous publications [2],[3], we have developed microfluidic platforms to enhance assessment and selection methods using electrical signal acquisition for sperm analysis. In addition, recent experiments showed that the presence of an acrosome on the sperm’s head can be measured by electrical opacity (impedance magnitude at high frequency divided by the magnitude at low frequency) [4]. Which leads to the question: what other characteristics of spermatozoa can be resolved with microfluidic impedance cytometry?
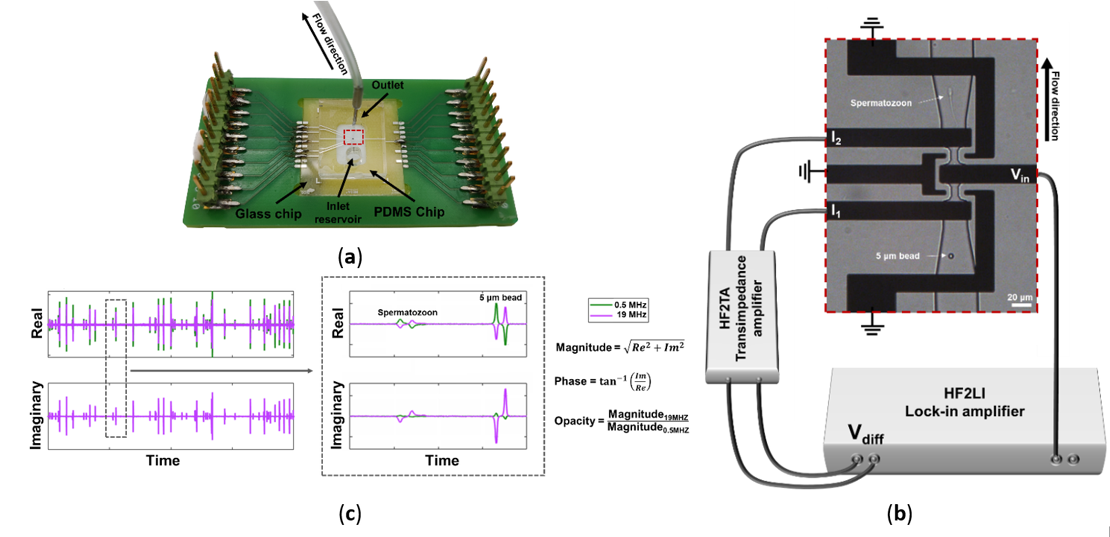
Figure 1. (a) Microfluidic impedance chip consisting of a PCB, glass chip and PDMS chip. (b) Microscopic image of electrodes and channels connected to the impedance spectroscope. An AC voltage is applied to the middle electrode (Vin), resulting in two currents at the two sensing electrodes (I1 and I2), which were then measured differentially (Vdiff). (c) Example of the real and imaginary signal of a passing bead and spermatozoon, respectively. The data were used to calculate the magnitude, phase and opacity of each event. [4]
Contact person: Stella Kruit (s.a.kruit@utwente.nl)
References
[1] Waberski, D., et al., Theriogenology 137 (2019): 2-7. DOI: 10.1016/j.theriogenology.2019.05.030
[2] Segerink, L. I., et al., 23.3 (2012): 66.
[3] de Wagenaar, B., et al., Lab on a Chip 16.8 (2016): 1514. DOI: 10.1039/C6LC00256K
[4] Kruit, S.A., et al., Biosensors 12.9 (2022): 679. DOI: 10.3390/bios12090679
