
PhD Student
E-mail: s.dewit@utwente.nl
Telephone: +31 53 489 4260
Fax: +31 53 489 3511
Address: Faculty of Science and Technology
Medical Cell BioPhysics (MCBP)
Room: CR4.435
Drienerlolaan 5
P.O. Box 217, 7500 AE Enschede
The Netherlands
Project goals
The CTCTrap project aims to isolate, identify and characterize all CTC in the blood of cancer patients. The capture and characterization of CTC enables a personalized approach in the diagnosis, prognosis and monitoring of cancer patients.
Project background
Circulating Tumor Cells (CTC) are cells that break free from the tumor and circulate in the blood of cancer patients, forming metastases. CTC provide real-time information about patient survival and treatment. Unfortunately, they are very rare and hard to capture and analyse. Currently under development is the CTCTrap, a therapeutic apheresis device that captures CTC from 5 litres blood from a metastatic cancer patient. A matrix captures CTC with antibodies and after release they are filtered through a sieve. Remaining cells are stained with antibodies and fluorescent molecules to discriminate between CTC and non-CTC. Subsequently, captured single CTC can be isolated and analysed. This will allow molecular characterization on single CTC, like protein, DNA and RNA analysis.
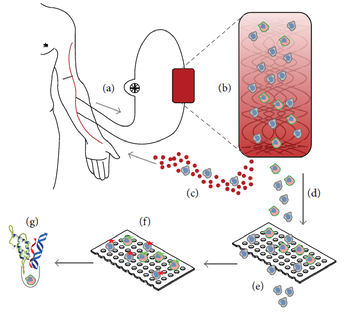
Figure 1 – Schematic representation of the CTCTrap. Blood from a patient (a) is passed through a functionalized 3D matrix (b). In this matrix are one or more specific antibodies present for CTC capture. A continuous blood flow without cells of interest is circled back to the patient (c). Captured cells are eluted from the matrix (d) and will be filtered through pores to reduce the background of cells that are not of interest (e). Cells on the filter can be used for immunofluorescent staining to discriminate CTC from blood cells (f) and subsequently be used for isolation of single CTC for additional molecular characterization, like protein, RNA and DNA analysis (g).
Molecular techniques for CTC characterization
The development of blood filtration with microsieves was used to detect CTC that are missed when the common epithelial marker EpCAM is not present on these cells. These cells are positive for the cytokeratin markers, showing it is not a cell of blood origin. The poster “EpCAM negative CTC in metastatic lung cancer enriched by filtration” describes the first results of a small study in lung cancer patients analysed with filtration. To show these captured cells are actually CTC, several molecular techniques are under development to perform on a microsieve; for instance fluorescent in situ hybridisation (FISH), comparative genomic hybridisation (CGH) and proximity ligation assay (PLA). Meanwhile, several clinical studies are running to determine the presence and clinical influence of these cells in lung- and prostate cancer.
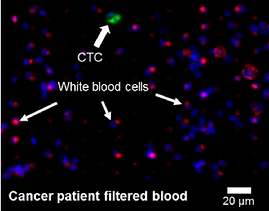
Figure 2 – The blood of cancer patients is filtered on a microsieve with pressure through 5 µm pores. Identification of cells is performed with fluorescent antibodies for the white blood cell marker CD45 (red) and the epithelial marker cytokeratin (green), combined with a nuclear DNA dye (blue).
